1. Conformational transitions of flexible macromolecules
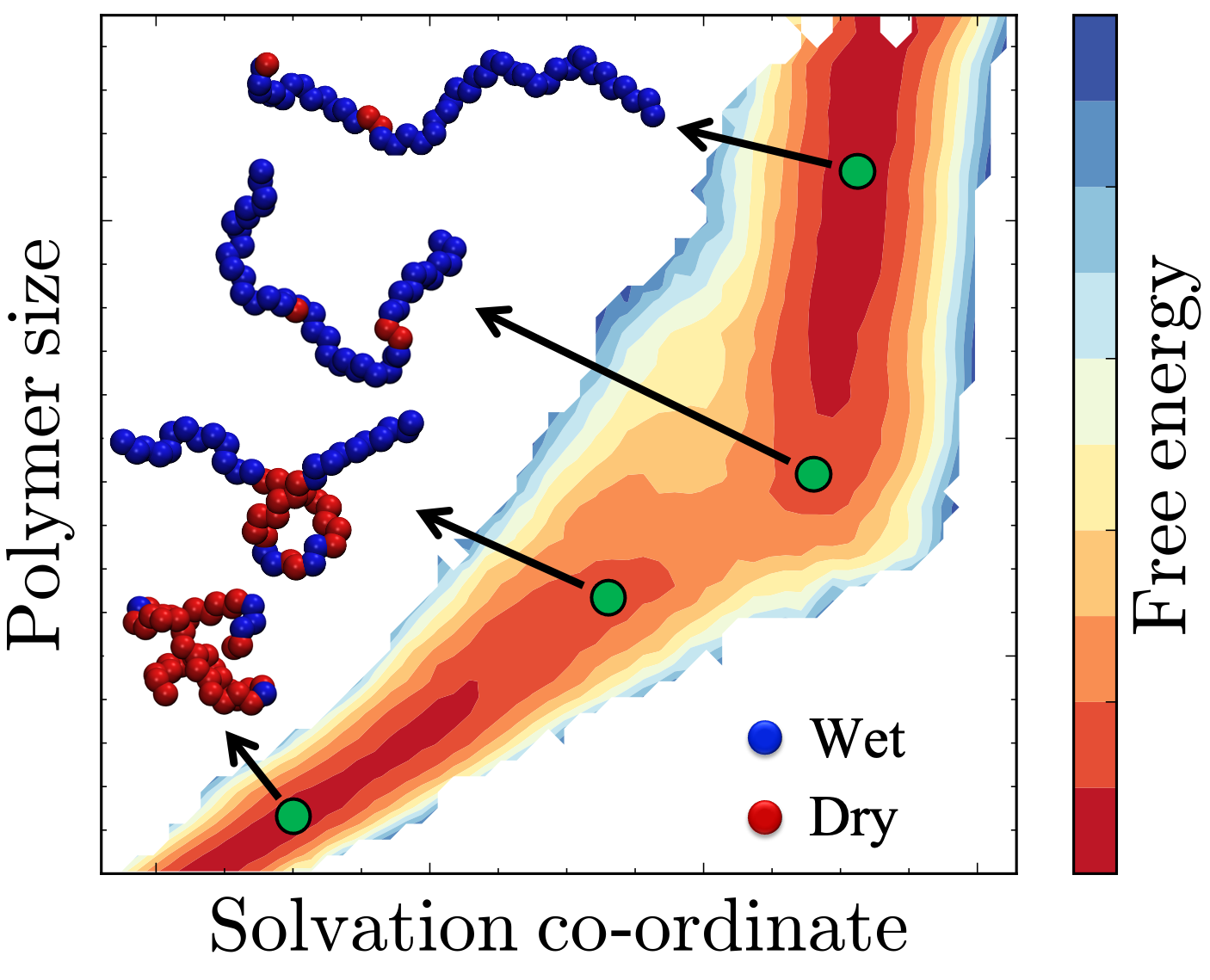
Flexible macromolecular solutes such as polymers, peptides, and proteins have the ability adopt a variety of distinct conformational states in solution. Polymers collapse into globular conformations in poor solvents and adopt extended conformations in good solvents or at favorable physical conditions. Peptides can fold to form defined secondary structures or adopt random coil configurations. Proteins can exist in their native folded states, transition to alternate folded conformations in response to binding events, post-translational modifications, or external forces, and partially or fully denature at low or high temperatures, high pressures, or high salt concentrations.
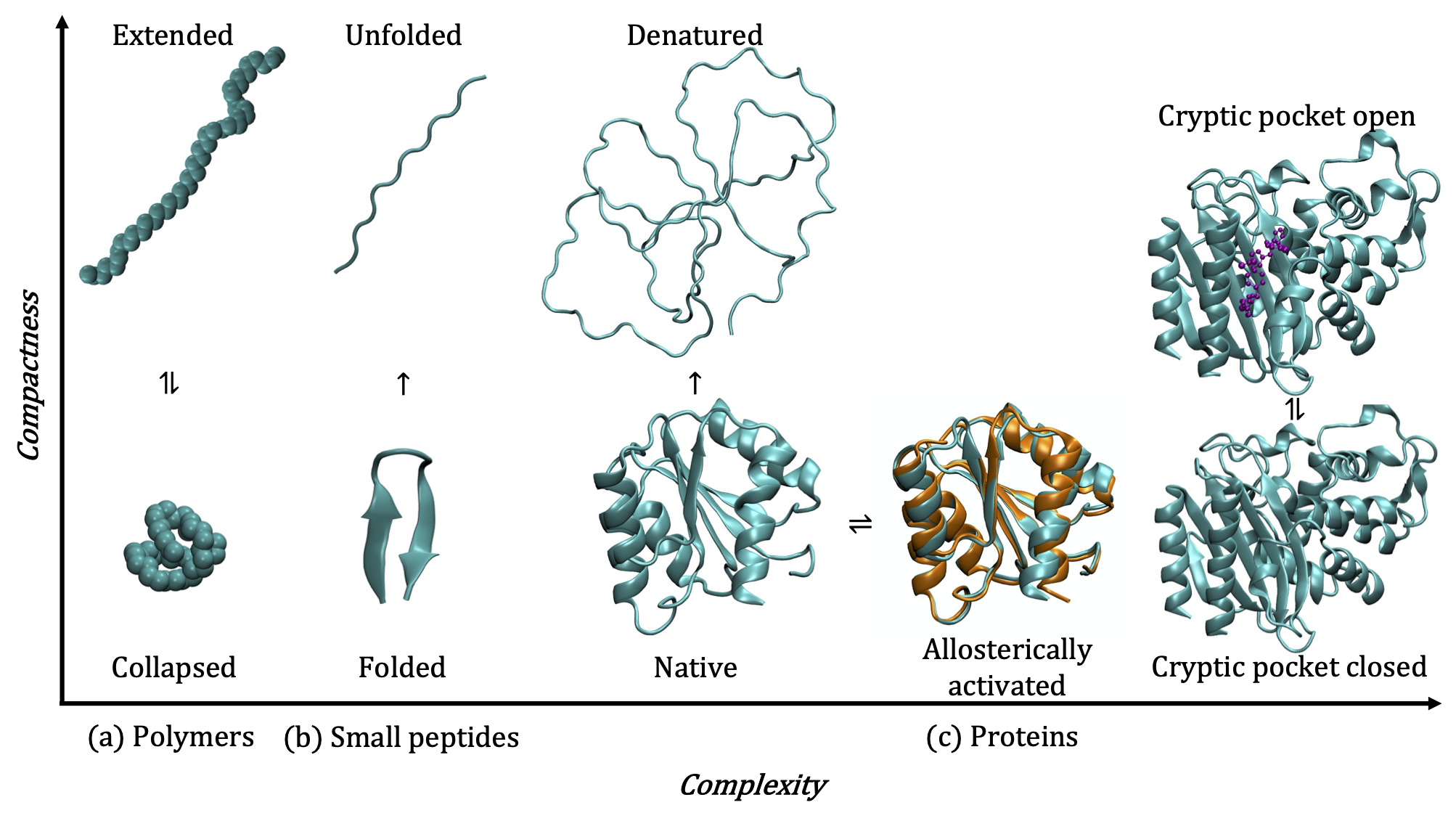
The ability to precisely modulate transitions between the myriad conformational states of proteins, peptides, and polymers has far-reaching applications in biology, ranging from the design of smart biomaterials to the design and delivery of drugs against specific protein targets. As these macromolecules are solvated in water in most biological contexts, their conformational transitions disrupt the inherent structure of water in many different ways, giving rise to rich phenomena such as the hydrophobic effect. Using state of the art molecular simulation and enhanced sampling techniques in conjuction with statistical thermodynamics and liquid state theory, my research focuses on characterizing the complex interplay between hydration and conformational transitions of proteins, peptides and polymers.
2. Decoding hydrophobicity with machine learning
Hydrophobicity plays a pivotal role in a myriad of biomolecular processes, spanning from intricate protein-ligand interactions to the orchestrated assembly of vast protein complexes. However, the hydrophobicity of diverse, chemically heterogeneous surfaces, such as those found on proteins, is not merely determined by their polar content but also by the nanoscale arrangements of polar and nonpolar groups adorning their surfaces. Consequently, traditional approaches like hydropathy scales fall short in accurately characterizing the hydrophobicity of these chemically diverse surfaces.
To confront the formidable challenge of predicting how surface chemical patterning influences hydrophobicity, my collaborative research endeavors harness the capabilities of machine learning. By comprehensively characterizing various measures of hydrophobicity using molecular simulation data derived from a diverse library of patterned surfaces, we aim to unravel the intricate interplay between surface chemistry and hydrophobic behavior, unlocking new frontiers in our understanding of this fundamental phenomenon.
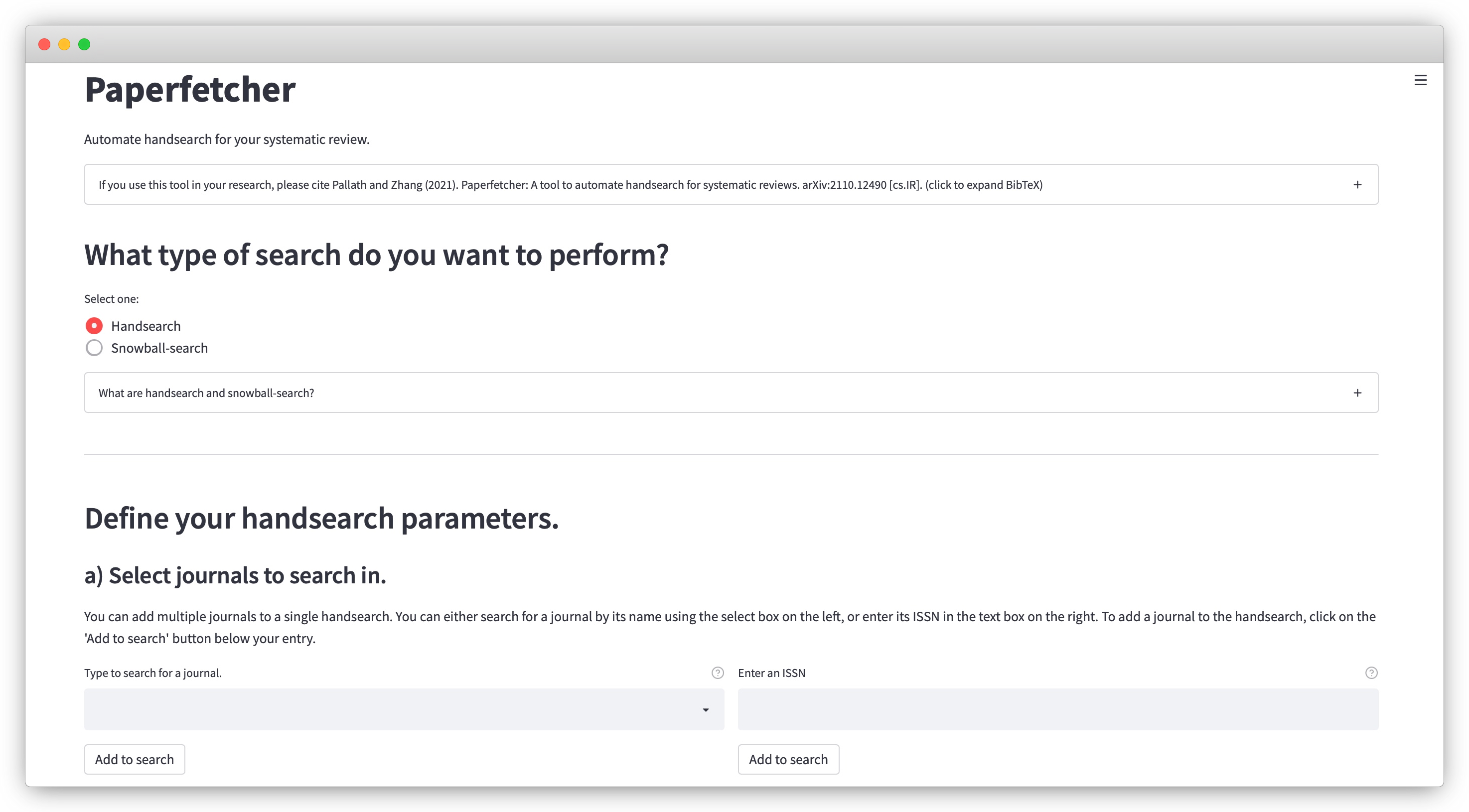
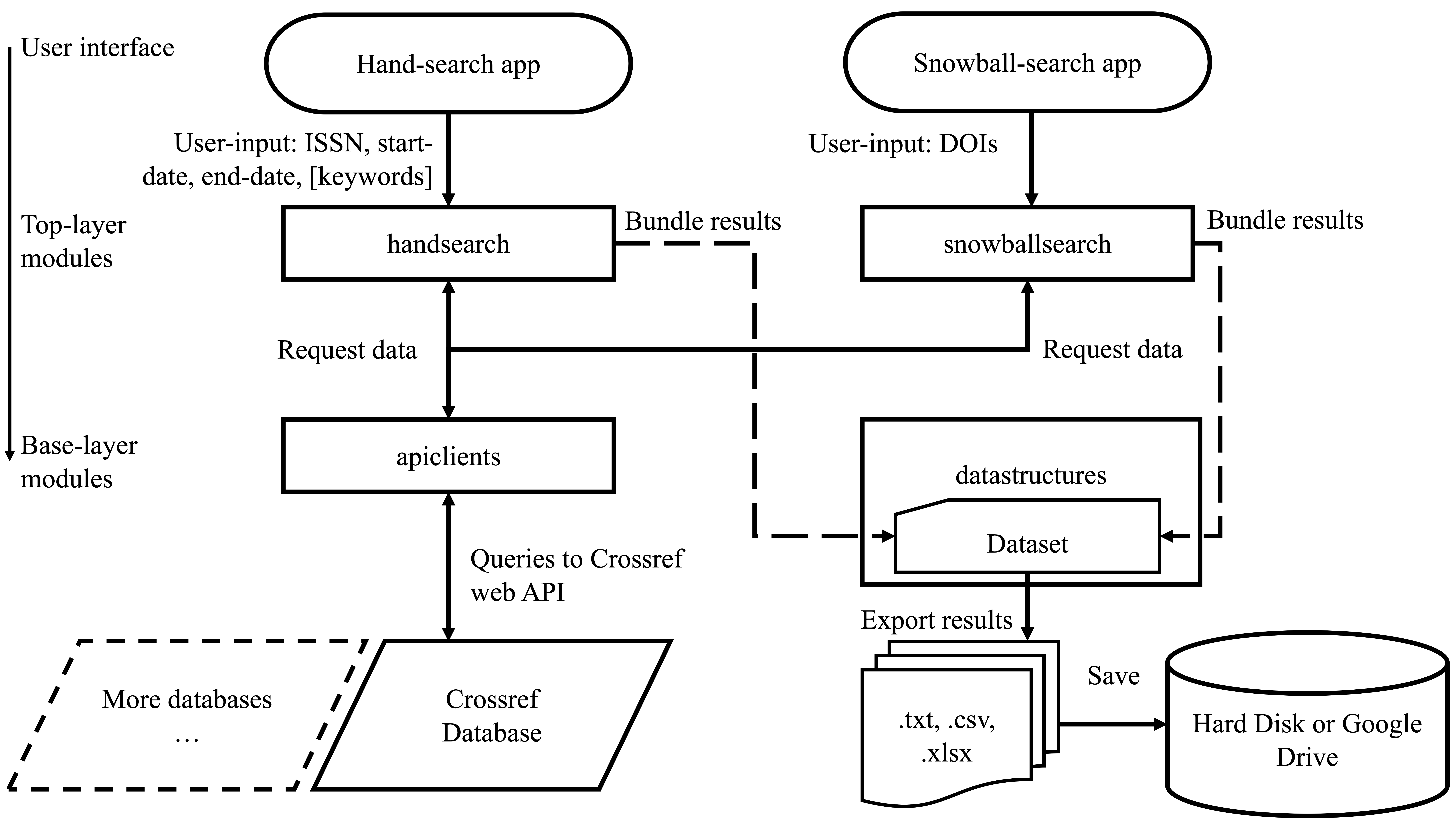
3. Tools for literature search
High-quality evidence synthesis relies on surveying and screening through hundreds to thousands of research articles. The process begins with literature search using electronic sources, which involves various complementary steps including bibliographic database search, handsearching, and citation searching. Some of these steps can be incredibly time-consuming, often requiring a team of researchers to invest several months of time for a single project. We are developing powerful cross-platform computational tools to help accelerate the process of literature search.
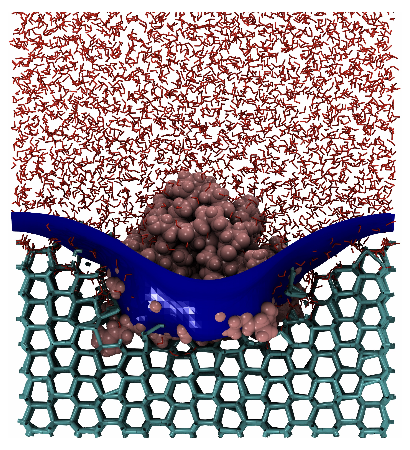
3 Thosar, A., Cai, Y., Marks, S. M., Vicars, Z., Choi, J., Pallath, A., & Patel, A. J. (2024). On the engulfment of antifreeze proteins by ice. Proceedings of the National Academy of Sciences.
Read on pnas.org >>
2 Pallath, A., & Zhang, Q. (2022). Paperfetcher: A tool to automate handsearching and citation searching for systematic reviews. Research Synthesis Methods. Software Focus.

Read on wiley.com >>
1 Dhabal, D., Jiang, Z., Pallath, A., & Patel, A. J. (2021). Characterizing the interplay between polymer solvation and conformation. The Journal of Physical Chemistry B, 125(20), 5434-5442. Published as part of the Lawrence R. Pratt Festschrift Virtual Special Issue.
Read on acs.org >>